• Soil microbes alter natural selection on plants facing competition and drought
How microorganisms affect the ecology and evolution of their larger plant and animal hosts is an exciting and important question. Our understanding of how animal health is influenced by their gut microbial communities has grown rapidly yet how soil and root microbiota influence plant health is still unclear. In order for host-associated microbes to affect host evolution three conditions need to be met: 1) the microbes associated with hosts must vary across individuals; 2) this variation must be heritable; and 3) there must be fitness differences associated with this variation across individual hosts.
We performed a large experiment examining how soil microorganisms alter the natural selection on plants facing competition and drought stress. We used natural accessions of the model plant Arabidopsis thaliana to explore whether soil microbes may be driving plant evolutionary responses to competition and drought.
Publications:
Fitzpatrick CR, Zainab M, Viliunas J. 2019. Soil microbes alter the natural selection of plants experiencing competition and drought. In Review.


• Phylogenetic relatedness, phenotypic similarity and plant-soil feedback
What determines how strongly species will interact with one another? Let’s say we’re talking about two species that occupy the same trophic level (i.e. they aren’t eating each other), if they share the same habitat and require the same resources then their interaction strength might be high. Identifying the species attributes that dictate ecological similarity is surprisingly difficult and as a proxy biologists have begun using the evolutionary relatedness between species to predict their interaction strength.
In this project we investigated how evolutionary relatedness and phenotypic similarity across 450 unique pairs of plant species predicts the outcome of their plant-soil feedback (PSF). PSF occurs when plants influence the abiotic/ biotic soil environment, which then influences the growth of another plant. PSF are important for numerous fundamental processes in terrestrial ecology (e.g. maintenance of plant diversity and productivity) and we’ve known about PSF for centuries – traditional agricultural practices like crop rotation are based on the PSF concept. Due to the unique evolutionary history of individual traits and the variability in their importance across interacting species, we show that indices of overall phenotypic and phylogenetic relatedness are poor predictors of plant-soil feedbacks at large phylogenetic scales.
Publications:
Fitzpatrick CR, Gehant L, Kotanen PM, Johnson MTJ. 2016. Phylogenetic relatedness, phenotypic similarity, and plant-soil feedbacks. Journal of Ecology 105: 786-800. PDF | Resources

• Genetic variation and contemporary evolution within plant species alters soil ecosystems
Individuals within a species can vary in ecologically important traits. For example, individuals can differ in their resource acquisition strategies, behaviour, morphology, and internal physiology. These individual differences can add up to affect population, community, and ecosystem-level processes. We think understanding these extended effects might be particularly important in scenarios where genetic variation is altered.
Individuals are usually arranged non-randomly across their range. Due to environmental gradients occurring at broad spatial scales, southern populations will exhibit phenotypic differences from northern populations. With global climate change species’ ranges are changing and populations are moving, sometimes rapidly, causing a large-scale reshuffling of genetic variation. Whether this introduction of novel genotypes will have consequences for associated communities and ecosystems is unknown. We’re testing whether the extended effects of genetic variation across the entire range of a focal plant species exhibit any geographical pattern. Specifically we’re looking at the effects of genetic variation in the plant species, Oenothera biennis, on the belowground invertebrate communities.

Publications:
Fitzpatrick CR, Mikhailitchenko A, Anstett D, Johnson MTJ. 2018. Range wide effect of plant genetic variation on soil invertebrate communities. Ecography 41: 1135-1146 . PDF
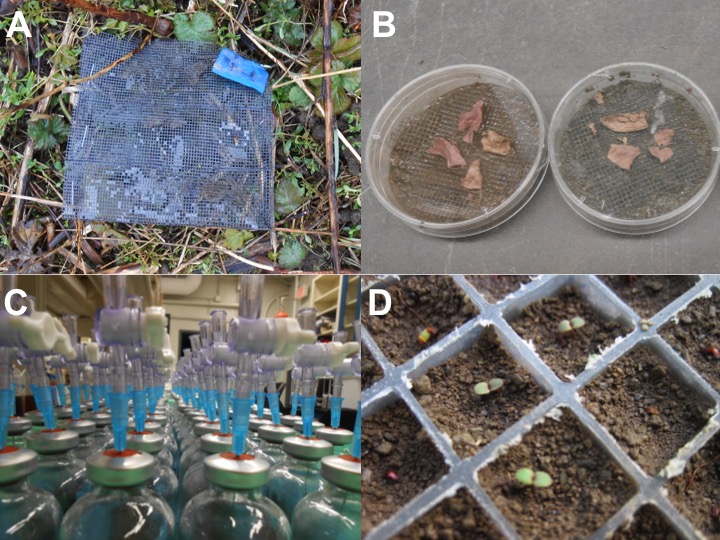
Evolution is another scenario where understanding the extended effects of genetic variation might be particularly important. When populations evolve accompanying shifts occur in the genetic variation present within the population. We wanted to know whether rapid evolution (<10 generations) could be driving ecosystem-level changes. We used replicate populations of the focal plant, Oenothera biennis, and manipulated the presence of insect herbivores on half the populations . After 5 generations of natural selection caused by insect herbivores evolution occurring within these populations actually influenced the rate of litter decomposition and nitrogen cycling. Additionally, we found that the ecosystem effects of evolution influenced the growth of plants in future generations. We were pretty excited by these results because they provide evidence for feedback loops between ecology and evolution, which to date has been very difficult to find.
Publications:
Fitzpatrick CR, Agrawal AA, Basiliko N, Hastings AP, Isaac ME, Preston M, Johnson MTJ. 2015. The importance of plant genotype and contemporary evolution for terrestrial ecosystem processes. Ecology 10: 2632-2642. PDF